Accelerating Data Analytics with Python and IBM dashDB
DashDB Show & Tell
Edouard Fouché
IBM Böblingen
26/09/2016


About me¶

Edouard Fouché¶
- Graduating from M. Sc. Computer Science at KIT
- Working student @ IBM Böblingen
- Developing Python tools for in-database analytics
- Interested in Data Mining & Cloud Computing
Outline¶
- Motivations
- Python for data science
- Limitations
- Bringing analytics to the data
- IBM dashDB
- Python/SQL
- Ibmdbpy
- Demo
- Possible extensions
- Conclusion/Discussion
Motivations¶
- The Python ecosystem for Data Science is rich
- Scipy, Numpy, Pandas
- Scikit-learn
- Matplotlib, Seaborn, Bokeh
- Jupyter notebooks...

Performance Limitation¶
- CPU, RAM
- Laptop / Workstation
Data Extraction¶
- Data is typically stored in data warehouses
- Big data volumes are impractical to extract
- What if the data is sensitive?
In-Database computation¶
Advantageous for several reasons:
- Databases are everywhere !
- Efficiency and scalability
- Data freshness
- Security

- Cloud-based data warehousing system
- Optimized for analytics and data mining
- Integrates BLU technology
- Data compression
- In-memory column store
- Massive Parallel Processing (MPP) scale-out

Example: Compute the mean for each column by class in the IRIS data set
SELECT AVG(PETALLENGTH), AVG(PETALWIDTH), AVG(SEPALLENGTH), AVG(SEPALWIDTH) FROM IRIS GROUP BY SPECIES
- Only one line in Python, using Pandas
In [1]:
from ibmdbpy.sampledata import iris
In [2]:
iris.groupby("species").mean()
Out[2]:
| sepal_length | sepal_width | petal_length | petal_width | |
|---|---|---|---|---|
| species | ||||
| setosa | 5.006 | 3.428 | 1.462 | 0.246 |
| versicolor | 5.936 | 2.770 | 4.260 | 1.326 |
| virginica | 6.588 | 2.974 | 5.552 | 2.026 |

The SQL-Pushdown approach¶
We translate higher level syntax into SQL
We push them to the underlying database
Everything happens transparently
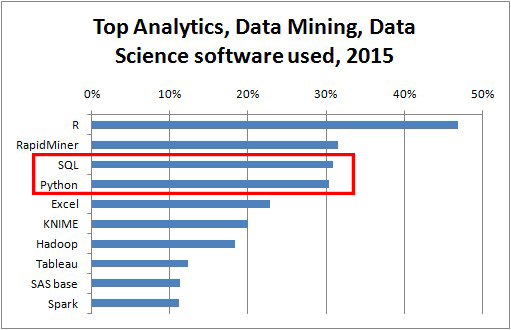
source: kdnuggets.com
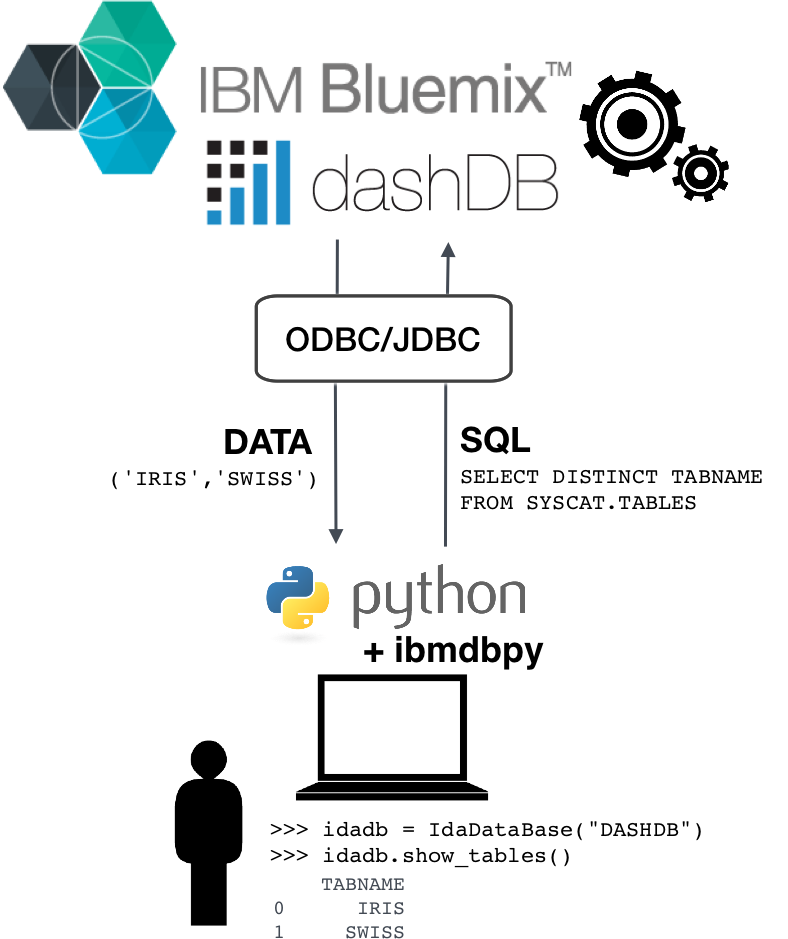
ibmdbpy¶
- Pandas-like interface for IBM dashDB
- Compatible from Python 2.7 up to 3.5
- ODBC or JDBC connection (cross-platform)
Provides high-level Python methods for:
- Database administration
- In-Database analytics
Demo¶
1: Connect to the database¶
In [4]:
from ibmdbpy import IdaDataBase
idadb = IdaDataBase("DASHDB", verbose=True)
IdaDataBase: abstraction layer for the distant database
- IDA : In-Database-Analytics
- Database administration
- Database lookup
- Upload Pandas.DataFrame as Table
- Download Table as Pandas.DataFrame
- Drop tables, create views...
2: Open a pointer to a table¶
In [5]:
from ibmdbpy import IdaDataFrame
iris = IdaDataFrame(idadb, "IRIS", indexer = "ID") # IRIS dataset
> SELECT distinct TABSCHEMA, TABNAME, OWNER, TYPE from SYSCAT.TABLES WHERE (OWNERTYPE = 'U') ORDER BY "TABSCHEMA","TABNAME" > SELECT TRIM(CURRENT_SCHEMA) FROM SYSIBM.SYSDUMMY1
- An IdaDataFrame instance is a pointer to a table in the database
- Data Manipulation
- Non-destructive data manipulation
- Statistics, filtering, sorting...
- Pandas-like syntax
3: Conduct data analytics¶
In [6]:
iris.head()
> SELECT * FROM DB2INST1.IRIS ORDER BY "ID" ASC FETCH FIRST 5 ROWS ONLY
Out[6]:
| ID | SEPALLENGTH | SEPALWIDTH | PETALLENGTH | PETALWIDTH | CLASS | |
|---|---|---|---|---|---|---|
| 0 | 1 | 5.1 | 3.5 | 1.4 | 0.2 | setosa |
| 1 | 2 | 4.9 | 3.0 | 1.4 | 0.2 | setosa |
| 2 | 3 | 4.7 | 3.2 | 1.3 | 0.2 | setosa |
| 3 | 4 | 4.6 | 3.1 | 1.5 | 0.2 | setosa |
| 4 | 5 | 5.0 | 3.6 | 1.4 | 0.2 | setosa |
Simple statistics¶
In [7]:
iris.corr()
> SELECT COLNAME, TYPENAME FROM SYSCAT.COLUMNS WHERE TABNAME='IRIS' AND TABSCHEMA='DB2INST1' ORDER BY COLNO
> SELECT CORRELATION("SEPALLENGTH","SEPALWIDTH"), CORRELATION("SEPALLENGTH","PETALLENGTH"), CORRELATION("SEPALLENGTH","PETALWIDTH"), CORRELATION("SEPALWIDTH","PETALLENGTH"), CORRELATION("SEPALWIDTH","PETALWIDTH"), CORRELATION("PETALLENGTH","PETALWIDTH") FROM DB2INST1.IRIS
Execution time: 0.21002697944641113 seconds.
Out[7]:
| SEPALLENGTH | SEPALWIDTH | PETALLENGTH | PETALWIDTH | |
|---|---|---|---|---|
| SEPALLENGTH | 1.000000 | -0.109369 | 0.871754 | 0.817954 |
| SEPALWIDTH | -0.109369 | 1.000000 | -0.420516 | -0.356544 |
| PETALLENGTH | 0.871754 | -0.420516 | 1.000000 | 0.962757 |
| PETALWIDTH | 0.817954 | -0.356544 | 0.962757 | 1.000000 |
In [8]:
iris.describe()
> SELECT COUNT("ID"), COUNT("SEPALLENGTH"), COUNT("SEPALWIDTH"), COUNT("PETALLENGTH"), COUNT("PETALWIDTH") FROM DB2INST1.IRIS
> SELECT AVG(CAST("ID" AS FLOAT)), AVG(CAST("SEPALLENGTH" AS FLOAT)), AVG(CAST("SEPALWIDTH" AS FLOAT)), AVG(CAST("PETALLENGTH" AS FLOAT)), AVG(CAST("PETALWIDTH" AS FLOAT)) FROM DB2INST1.IRIS
> SELECT COUNT("ID"), COUNT("SEPALLENGTH"), COUNT("SEPALWIDTH"), COUNT("PETALLENGTH"), COUNT("PETALWIDTH") FROM DB2INST1.IRIS
> SELECT STDDEV("ID")*(SQRT(150.0)/SQRT(149.0)), STDDEV("SEPALLENGTH")*(SQRT(150.0)/SQRT(149.0)), STDDEV("SEPALWIDTH")*(SQRT(150.0)/SQRT(149.0)), STDDEV("PETALLENGTH")*(SQRT(150.0)/SQRT(149.0)), STDDEV("PETALWIDTH")*(SQRT(150.0)/SQRT(149.0)) FROM DB2INST1.IRIS
> SELECT MIN("ID"), MIN("SEPALLENGTH"), MIN("SEPALWIDTH"), MIN("PETALLENGTH"), MIN("PETALWIDTH") FROM DB2INST1.IRIS
> SELECT * FROM (SELECT COUNT(*) AS "ID" FROM DB2INST1.IRIS WHERE "ID" IS NULL), (SELECT COUNT(*) AS "SEPALLENGTH" FROM DB2INST1.IRIS WHERE "SEPALLENGTH" IS NULL), (SELECT COUNT(*) AS "SEPALWIDTH" FROM DB2INST1.IRIS WHERE "SEPALWIDTH" IS NULL), (SELECT COUNT(*) AS "PETALLENGTH" FROM DB2INST1.IRIS WHERE "PETALLENGTH" IS NULL), (SELECT COUNT(*) AS "PETALWIDTH" FROM DB2INST1.IRIS WHERE "PETALWIDTH" IS NULL)
> SELECT CAST(COUNT(*) AS INTEGER) FROM DB2INST1.IRIS
> (SELECT "ID" AS "ID" FROM (SELECT ROW_NUMBER() OVER(ORDER BY "ID") as rn, "ID" FROM (SELECT * FROM DB2INST1.IRIS)) WHERE rn in(38,39,75,76,112,113))
> (SELECT "SEPALLENGTH" AS "SEPALLENGTH" FROM (SELECT ROW_NUMBER() OVER(ORDER BY "SEPALLENGTH") as rn, "SEPALLENGTH" FROM (SELECT * FROM DB2INST1.IRIS)) WHERE rn in(38,39,75,76,112,113))
> (SELECT "SEPALWIDTH" AS "SEPALWIDTH" FROM (SELECT ROW_NUMBER() OVER(ORDER BY "SEPALWIDTH") as rn, "SEPALWIDTH" FROM (SELECT * FROM DB2INST1.IRIS)) WHERE rn in(38,39,75,76,112,113))
> (SELECT "PETALLENGTH" AS "PETALLENGTH" FROM (SELECT ROW_NUMBER() OVER(ORDER BY "PETALLENGTH") as rn, "PETALLENGTH" FROM (SELECT * FROM DB2INST1.IRIS)) WHERE rn in(38,39,75,76,112,113))
> (SELECT "PETALWIDTH" AS "PETALWIDTH" FROM (SELECT ROW_NUMBER() OVER(ORDER BY "PETALWIDTH") as rn, "PETALWIDTH" FROM (SELECT * FROM DB2INST1.IRIS)) WHERE rn in(38,39,75,76,112,113))
> SELECT MAX("ID"), MAX("SEPALLENGTH"), MAX("SEPALWIDTH"), MAX("PETALLENGTH"), MAX("PETALWIDTH") FROM DB2INST1.IRIS
Execution time: 1.752722978591919 seconds.
Out[8]:
| ID | SEPALLENGTH | SEPALWIDTH | PETALLENGTH | PETALWIDTH | |
|---|---|---|---|---|---|
| count | 150.000000 | 150.000000 | 150.000000 | 150.000000 | 150.000000 |
| mean | 75.500000 | 5.843333 | 3.054000 | 3.758667 | 1.198667 |
| std | 43.445368 | 0.828066 | 0.433594 | 1.764420 | 0.763161 |
| min | 1.000000 | 4.300000 | 2.000000 | 1.000000 | 0.100000 |
| 25% | 38.500000 | 5.100000 | 2.800000 | 1.600000 | 0.300000 |
| 50% | 75.500000 | 5.800000 | 3.000000 | 4.350000 | 1.300000 |
| 75% | 112.500000 | 6.400000 | 3.300000 | 5.100000 | 1.800000 |
| max | 150.000000 | 7.900000 | 4.400000 | 6.900000 | 2.500000 |
Data manipulation¶
In [9]:
iris = iris[["ID","SEPALLENGTH", "SEPALWIDTH"]]
In [10]:
iris['new'] = iris['SEPALLENGTH']**2 + iris['SEPALWIDTH'].mean()
> CREATE VIEW "TEMP_VIEW_IRIS_89613_1474908989" AS (SELECT "SEPALLENGTH" FROM DB2INST1.IRIS)
<< AUTOCOMMIT >>
> SELECT COLNAME, TYPENAME FROM SYSCAT.COLUMNS WHERE TABNAME='TEMP_VIEW_IRIS_89613_1474908989' ORDER BY COLNO
> DROP VIEW "TEMP_VIEW_IRIS_89613_1474908989"
<< AUTOCOMMIT >>
> CREATE VIEW "TEMP_VIEW_IRIS_13064_1474908989" AS (SELECT "SEPALWIDTH" FROM DB2INST1.IRIS)
<< AUTOCOMMIT >>
> SELECT COLNAME, TYPENAME FROM SYSCAT.COLUMNS WHERE TABNAME='TEMP_VIEW_IRIS_13064_1474908989' ORDER BY COLNO
> DROP VIEW "TEMP_VIEW_IRIS_13064_1474908989"
<< AUTOCOMMIT >>
> SELECT AVG(CAST("SEPALWIDTH" AS FLOAT)) FROM (SELECT "SEPALWIDTH" FROM DB2INST1.IRIS)
Execution time: 0.26253414154052734 seconds.
> CREATE VIEW "TEMP_VIEW_IRIS_71822_1474908990" AS (SELECT ( POWER("SEPALLENGTH",2)) AS "SEPALLENGTH" FROM DB2INST1.IRIS)
<< AUTOCOMMIT >>
> SELECT COLNAME, TYPENAME FROM SYSCAT.COLUMNS WHERE TABNAME='TEMP_VIEW_IRIS_71822_1474908990' ORDER BY COLNO
> DROP VIEW "TEMP_VIEW_IRIS_71822_1474908990"
<< AUTOCOMMIT >>
In [11]:
iris.head()
> SELECT * FROM (SELECT "ID","SEPALLENGTH","SEPALWIDTH",(( POWER("SEPALLENGTH",2)) + 3.054) AS "new" FROM DB2INST1.IRIS) ORDER BY "ID" ASC FETCH FIRST 5 ROWS ONLY
Out[11]:
| ID | SEPALLENGTH | SEPALWIDTH | new | |
|---|---|---|---|---|
| 0 | 1 | 5.1 | 3.5 | 29.064 |
| 1 | 2 | 4.9 | 3.0 | 27.064 |
| 2 | 3 | 4.7 | 3.2 | 25.144 |
| 3 | 4 | 4.6 | 3.1 | 24.214 |
| 4 | 5 | 5.0 | 3.6 | 28.054 |
Remark: Data stays in the database¶
In [12]:
iris
Out[12]:
<ibmdbpy.frame.IdaDataFrame at 0xa97f208>
In [13]:
iris.print()
SELECT "ID","SEPALLENGTH","SEPALWIDTH",(( POWER("SEPALLENGTH",2)) + 3.054) AS "new" FROM DB2INST1.IRIS
Machine Learning¶
- IBM dashDB is more than just a database
- Includes in-database Data Mining algorithms
- Currently available in ibmdbpy :
- K-means
- Naive Bayes
- Association Rules
In [14]:
from ibmdbpy.learn import KMeans
kmeans = KMeans(3) # clustering with 3 clusters
In [15]:
iris = IdaDataFrame(idadb, "IRIS", indexer = "ID")
In [16]:
kmeans.fit_predict(iris).head()
> SELECT COUNT(*) FROM SYSCAT.ROUTINES WHERE ROUTINENAME='KMEANS' AND ROUTINEMODULENAME = 'IDAX'
> CALL IDAX.KMEANS ('model="KMEANS_44003_1474908991",distance="euclidean",randseed=12345,intable=IRIS,k=3,idbased=False,id="ID",maxiter=5')
<< AUTOCOMMIT >>
<< COMMIT >>
> SELECT * FROM "DB2INST1"."KMEANS_44003_1474908991_MODEL"
> SELECT * FROM "DB2INST1"."KMEANS_44003_1474908991_COLUMNS"
> SELECT * FROM "DB2INST1"."KMEANS_44003_1474908991_COLUMN_STATISTICS"
> SELECT * FROM "DB2INST1"."KMEANS_44003_1474908991_CLUSTERS"
> CALL IDAX.PREDICT_KMEANS ('model="KMEANS_44003_1474908991",outtable="PREDICT_KMEANS_62820_1474909004",id="ID",intable=IRIS')
<< AUTOCOMMIT >>
<< COMMIT >>
> SELECT distinct TABSCHEMA, TABNAME, OWNER, TYPE from SYSCAT.TABLES WHERE (OWNERTYPE = 'U') ORDER BY "TABSCHEMA","TABNAME"
> SELECT * FROM DB2INST1.PREDICT_KMEANS_62820_1474909004 ORDER BY "ID" ASC FETCH FIRST 5 ROWS ONLY
Out[16]:
| ID | CLUSTER_ID | DISTANCE | |
|---|---|---|---|
| 0 | 1 | 1 | 0.332119 |
| 1 | 2 | 2 | 0.228125 |
| 2 | 3 | 2 | 0.138505 |
| 3 | 4 | 2 | 0.141902 |
| 4 | 5 | 1 | 0.382005 |
In [17]:
kmeans.describe()
KMeans clustering with 3 clusters of sizes 32, 97, 21
Cluster means:
CLUSTERID SEPALLENGTH SEPALWIDTH PETALLENGTH PETALWIDTH CLASS
0 1 5.209375 3.531250 1.671875 0.353125 setosa
1 2 4.695238 3.100000 1.395238 0.190476 setosa
2 3 6.301031 2.886598 4.958763 1.695876 virginica
Within cluster sum of squares by cluster:
[ 22.02570934 170.84912849 2.75907407]
> CALL IDAX.PRINT_MODEL ('model="KMEANS_44003_1474908991"')
<< AUTOCOMMIT >>
In-database Feature Selection¶
- Master Thesis: Fast In-Database Feature Selection
- Mapping feature selection algorithm to databases
- Outcome: Efficient approaches for various correlation matrices
- Pearson
- Chi-squared
- Gini index
- Mutual information
- Gain ratio
- and more...
In [18]:
iris = IdaDataFrame(idadb, "IRIS_DISC", indexer="ID")
In [19]:
from ibmdbpy.feature_selection import info_gain
info_gain(iris)
> SELECT CAST(COUNT(*) AS INTEGER) FROM DB2INST1.IRIS_DISC > SELECT SUM(-a*LOG(a)) FROM(SELECT COUNT(*) AS a FROM DB2INST1.IRIS_DISC GROUP BY "SEPALLENGTH") > SELECT SUM(-a*LOG(a)) FROM(SELECT COUNT(*) AS a FROM DB2INST1.IRIS_DISC GROUP BY "SEPALWIDTH") > SELECT SUM(-a*LOG(a)) FROM(SELECT COUNT(*) AS a FROM DB2INST1.IRIS_DISC GROUP BY "SEPALLENGTH","SEPALWIDTH") > SELECT SUM(-a*LOG(a)) FROM(SELECT COUNT(*) AS a FROM DB2INST1.IRIS_DISC GROUP BY "PETALLENGTH") > SELECT SUM(-a*LOG(a)) FROM(SELECT COUNT(*) AS a FROM DB2INST1.IRIS_DISC GROUP BY "SEPALLENGTH","PETALLENGTH") > SELECT SUM(-a*LOG(a)) FROM(SELECT COUNT(*) AS a FROM DB2INST1.IRIS_DISC GROUP BY "PETALWIDTH") > SELECT SUM(-a*LOG(a)) FROM(SELECT COUNT(*) AS a FROM DB2INST1.IRIS_DISC GROUP BY "SEPALLENGTH","PETALWIDTH") > SELECT SUM(-a*LOG(a)) FROM(SELECT COUNT(*) AS a FROM DB2INST1.IRIS_DISC GROUP BY "CLASS") > SELECT SUM(-a*LOG(a)) FROM(SELECT COUNT(*) AS a FROM DB2INST1.IRIS_DISC GROUP BY "SEPALLENGTH","CLASS") > SELECT SUM(-a*LOG(a)) FROM(SELECT COUNT(*) AS a FROM DB2INST1.IRIS_DISC GROUP BY "SEPALWIDTH","PETALLENGTH") > SELECT SUM(-a*LOG(a)) FROM(SELECT COUNT(*) AS a FROM DB2INST1.IRIS_DISC GROUP BY "SEPALWIDTH","PETALWIDTH") > SELECT SUM(-a*LOG(a)) FROM(SELECT COUNT(*) AS a FROM DB2INST1.IRIS_DISC GROUP BY "SEPALWIDTH","CLASS") > SELECT SUM(-a*LOG(a)) FROM(SELECT COUNT(*) AS a FROM DB2INST1.IRIS_DISC GROUP BY "PETALLENGTH","PETALWIDTH") > SELECT SUM(-a*LOG(a)) FROM(SELECT COUNT(*) AS a FROM DB2INST1.IRIS_DISC GROUP BY "PETALLENGTH","CLASS") > SELECT SUM(-a*LOG(a)) FROM(SELECT COUNT(*) AS a FROM DB2INST1.IRIS_DISC GROUP BY "PETALWIDTH","CLASS") Execution time: 2.4338090419769287 seconds.
Out[19]:
| SEPALLENGTH | SEPALWIDTH | PETALLENGTH | PETALWIDTH | CLASS | |
|---|---|---|---|---|---|
| SEPALLENGTH | NaN | 0.163915 | 0.708674 | 0.675541 | 0.652284 |
| SEPALWIDTH | 0.163915 | NaN | 0.381823 | 0.397607 | 0.376050 |
| PETALLENGTH | 0.708674 | 0.381823 | NaN | 1.346137 | 1.356545 |
| PETALWIDTH | 0.675541 | 0.397607 | 1.346137 | NaN | 1.378403 |
| CLASS | 0.652284 | 0.376050 | 1.356545 | 1.378403 | NaN |
Performance comparison¶
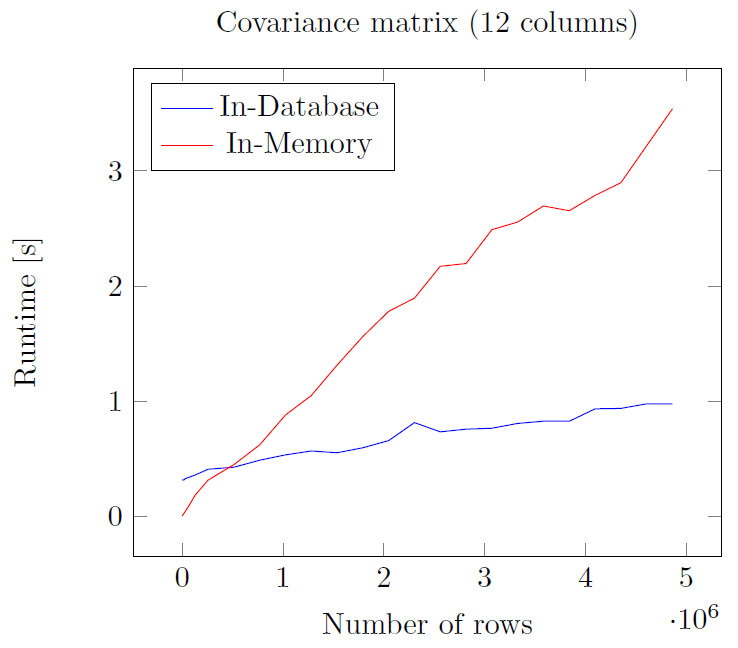
- In-Database: IBM dashDB entry plan
- In-Memory: Notebook, i5 2.6GHz - 16GB RAM
Deployment¶
- Distribution via PyPI
pip install ibmdbpy
- Available on GitHub: https://github.com/ibmdbanalytics/ibmdbpy
- License : BSD
- Version : 0.1.3 (September 2016)
- Documentation: http://pythonhosted.org/ibmdbpy/
Possible extensions¶
- New features:
- In-Database Feature Selection
- In-Database Geospatial Analytics
- More features (todo):
- Merge, sampling
- Grouping
- More ML wrappers
- Sequential patterns
- Linear regression
- Decisions trees, Regression trees
- k-NN
Conclusion¶
- Ibmdbpy is an interface for in-database computing
- Python / IBM dashDB
- No data extraction required
- Data freshness
- Security
- Intuitive: Pandas-like, Sklearn-like syntax
- Shows great performance on "big" datasets
- Nice tool for further applications: Feature Selection, Geospatial
- Drawbacks:
- Requires to connect to a remote database instance
- Designed only for IBM dashDB / IBM DB2
- Covers only a small part of Python analytics capabilities
- Note: For R users, use ibmdbR
Thank you for your attention !¶
Any questions, suggestions ?

