Accelerating Python Analytics by
In-Database Processing
PyData Berlin 2016
Edouard Fouché
IBM Research & Development
20/05/2016

About me¶

Edouard Fouché¶
- M.Eng. from ESIEE PARIS
- Graduate CS student at Karlsruhe Institute of Technology (KIT)
- Interested in Machine Learning & Cloud Computing
- Working student at IBM since April ’15
- Developing Python tools for in-database analytics
Outline¶
- Motivations
- Python for data science
- Limitations
- Solutions
- Bringing analytics to the data
- IBM dashDB
- Python/SQL
- Ibmdbpy
- Demo !
- Conclusion/Discussion
Motivations¶
- The Python ecosystem for Data Science is rich
- Scipy, Numpy, Pandas
- Scikit-learn
- Matplotlib, Seaborn, Bokeh
- IPython notebooks...

Performance Limitation¶
- CPU, RAM
- Laptop / Workstation
Data Extraction¶
- Data is typically stored in data warehouses
- Big data volumes are impractical to extract
Solutions¶
Out-of-core: Data does not fit in the main memory
- In-Workstation computation, e.g. Dask
- In-Cluster computation, e.g. Hadoop, Spark
- In-Database computation
- Directly in the database engine
In-Database computation¶
Advantageous for several reasons:
- Databases are everywhere !
- Efficiency and scalability
- Data freshness
- Security

- Cloud-based data warehousing system
- Optimized for analytics and data mining
- Integrates BLU technology
- Data compression
- In-memory column store
- Massive Parallel Processing (MPP) scale-out

Task: Compute the mean for each column given the class in the IRIS data set
SELECT AVG(PETALLENGTH), AVG(PETALWIDTH),
AVG(SEPALLENGTH), AVG(SEPALWIDTH)
FROM IRIS
GROUP BY SPECIES
- Only one line in Pandas
In [21]:
from ibmdbpy.sampledata import iris
In [22]:
iris.groupby("species").mean()
Out[22]:
sepal_length
sepal_width
petal_length
petal_width
species
setosa
5.006
3.428
1.462
0.246
versicolor
5.936
2.770
4.260
1.326
virginica
6.588
2.974
5.552
2.026

- Pandas-like interface for multiple backends
- Translates Python code into something else (e.g. SQL)
- Supports a lot of backends
However
- Only a subset of functions for each backend
- Cannot make use of backends' specific functions

The SQL-Pushdown approach¶
We translate higher level syntax into SQL
We push them to the underlying database
Everything happens transparently
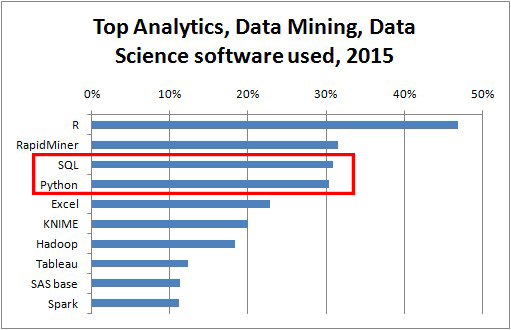 source : KDnuggets 2015
source : KDnuggets 2015
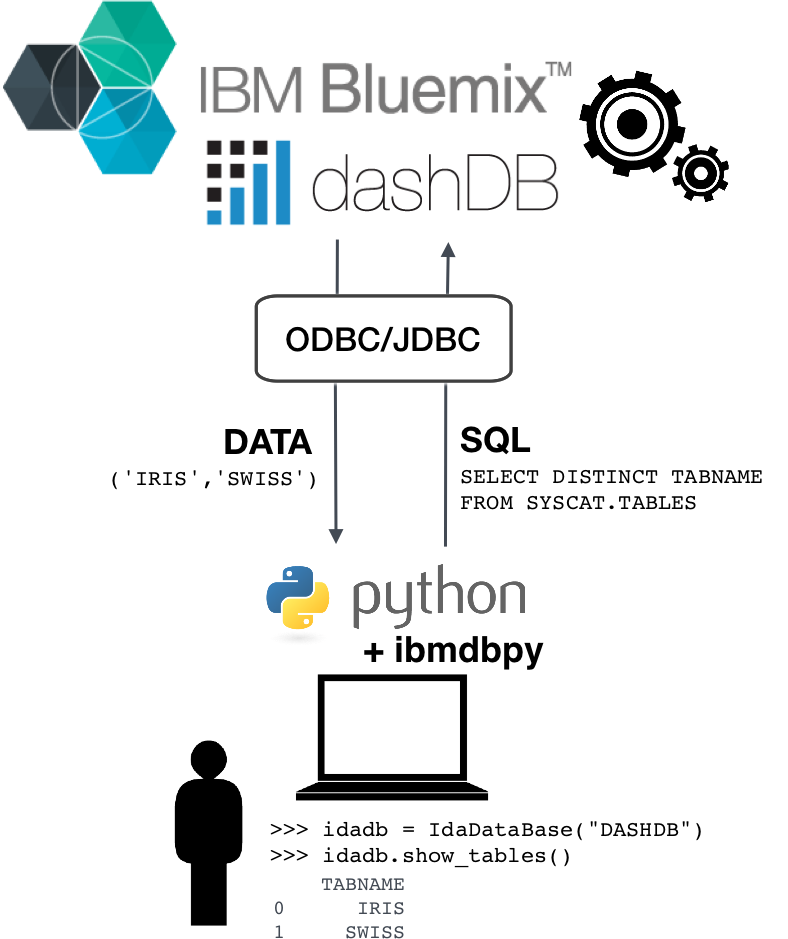
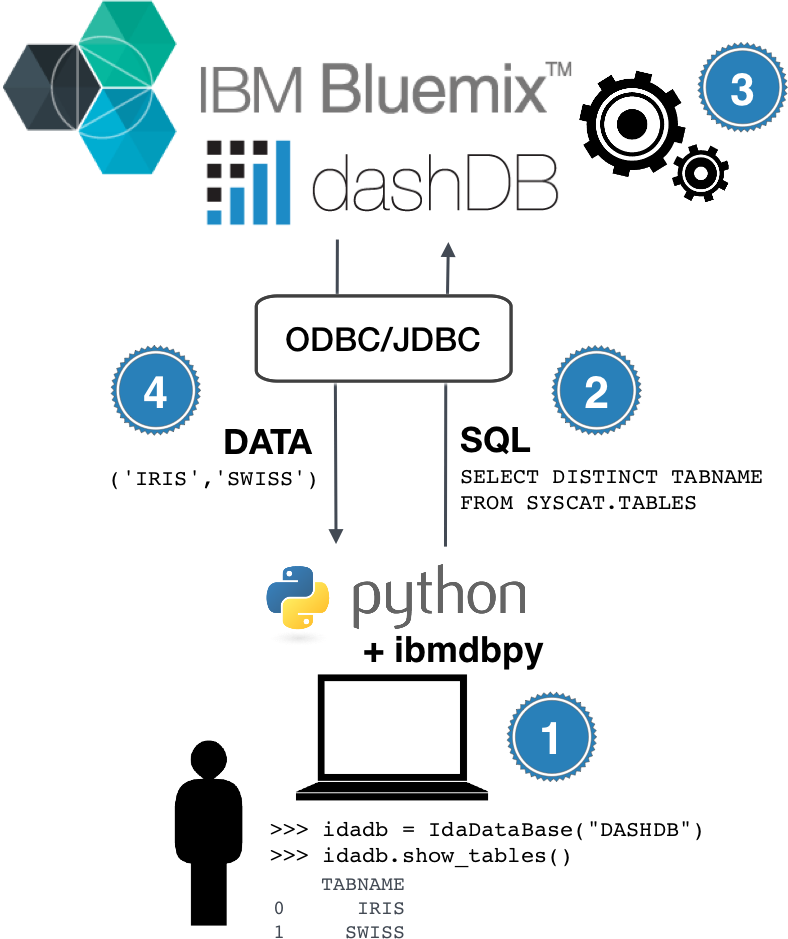
ibmdbpy¶
- Pandas-like interface for IBM dashDB
- Compatible Python 2.7 up to 3.5
- ODBC or JDBC connection (cross-platform)
- Data analytics
- Database administration
Demo¶
Connect to the database¶
In [23]:
from ibmdbpy import IdaDataBase
idadb = IdaDataBase("DASHDB", verbose=True)
IdaDataBase instances are an abstraction layer for the connection to dashDB
- IDA : In-Database-Analytics
- Database administration
- Database lookup
- Upload Pandas.DataFrame, Download Table
- Drop tables, create views...
Open a pointer to a table¶
In [24]:
from ibmdbpy import IdaDataFrame
iris = IdaDataFrame(idadb, "IRIS", indexer = "ID") # IRIS dataset
> SELECT distinct TABSCHEMA, TABNAME, OWNER, TYPE from SYSCAT.TABLES WHERE (OWNERTYPE = 'U') ORDER BY "TABSCHEMA","TABNAME"
> SELECT TRIM(CURRENT_SCHEMA) FROM SYSIBM.SYSDUMMY1
- An IdaDataFrame instance is a pointer to a table in the database
- Data Manipulation
- Non-destructive data manipulation
- Statistics, filtering, sorting...
- Pandas-like syntax
In [25]:
iris.head()
> SELECT * FROM DB2INST1.IRIS ORDER BY "ID" ASC FETCH FIRST 5 ROWS ONLY
Out[25]:
ID
SEPALLENGTH
SEPALWIDTH
PETALLENGTH
PETALWIDTH
CLASS
0
1
5.1
3.5
1.4
0.2
setosa
1
2
4.9
3.0
1.4
0.2
setosa
2
3
4.7
3.2
1.3
0.2
setosa
3
4
4.6
3.1
1.5
0.2
setosa
4
5
5.0
3.6
1.4
0.2
setosa
Simple statistics¶
In [26]:
iris.corr()
> SELECT COLNAME, TYPENAME FROM SYSCAT.COLUMNS WHERE TABNAME='IRIS' AND TABSCHEMA='DB2INST1' ORDER BY COLNO
> SELECT CORRELATION("SEPALLENGTH","SEPALWIDTH"), CORRELATION("SEPALLENGTH","PETALLENGTH"), CORRELATION("SEPALLENGTH","PETALWIDTH"), CORRELATION("SEPALWIDTH","PETALLENGTH"), CORRELATION("SEPALWIDTH","PETALWIDTH"), CORRELATION("PETALLENGTH","PETALWIDTH") FROM DB2INST1.IRIS
Execution time: 0.16952085494995117 seconds.
Out[26]:
SEPALLENGTH
SEPALWIDTH
PETALLENGTH
PETALWIDTH
SEPALLENGTH
1.000000
-0.109369
0.871754
0.817954
SEPALWIDTH
-0.109369
1.000000
-0.420516
-0.356544
PETALLENGTH
0.871754
-0.420516
1.000000
0.962757
PETALWIDTH
0.817954
-0.356544
0.962757
1.000000
In [27]:
iris.describe()
> SELECT COUNT("ID"), COUNT("SEPALLENGTH"), COUNT("SEPALWIDTH"), COUNT("PETALLENGTH"), COUNT("PETALWIDTH") FROM DB2INST1.IRIS
> SELECT AVG(CAST("ID" AS FLOAT)), AVG(CAST("SEPALLENGTH" AS FLOAT)), AVG(CAST("SEPALWIDTH" AS FLOAT)), AVG(CAST("PETALLENGTH" AS FLOAT)), AVG(CAST("PETALWIDTH" AS FLOAT)) FROM DB2INST1.IRIS
> SELECT COUNT("ID"), COUNT("SEPALLENGTH"), COUNT("SEPALWIDTH"), COUNT("PETALLENGTH"), COUNT("PETALWIDTH") FROM DB2INST1.IRIS
> SELECT STDDEV("ID")*(SQRT(150.0)/SQRT(149.0)), STDDEV("SEPALLENGTH")*(SQRT(150.0)/SQRT(149.0)), STDDEV("SEPALWIDTH")*(SQRT(150.0)/SQRT(149.0)), STDDEV("PETALLENGTH")*(SQRT(150.0)/SQRT(149.0)), STDDEV("PETALWIDTH")*(SQRT(150.0)/SQRT(149.0)) FROM DB2INST1.IRIS
> SELECT MIN("ID"), MIN("SEPALLENGTH"), MIN("SEPALWIDTH"), MIN("PETALLENGTH"), MIN("PETALWIDTH") FROM DB2INST1.IRIS
> SELECT * FROM (SELECT COUNT(*) AS "ID" FROM DB2INST1.IRIS WHERE "ID" IS NULL), (SELECT COUNT(*) AS "SEPALLENGTH" FROM DB2INST1.IRIS WHERE "SEPALLENGTH" IS NULL), (SELECT COUNT(*) AS "SEPALWIDTH" FROM DB2INST1.IRIS WHERE "SEPALWIDTH" IS NULL), (SELECT COUNT(*) AS "PETALLENGTH" FROM DB2INST1.IRIS WHERE "PETALLENGTH" IS NULL), (SELECT COUNT(*) AS "PETALWIDTH" FROM DB2INST1.IRIS WHERE "PETALWIDTH" IS NULL)
> SELECT CAST(COUNT(*) AS INTEGER) FROM DB2INST1.IRIS
> (SELECT "ID" AS "ID" FROM (SELECT ROW_NUMBER() OVER(ORDER BY "ID") as rn, "ID" FROM (SELECT * FROM DB2INST1.IRIS)) WHERE rn in(38,39,75,76,112,113))
> (SELECT "SEPALLENGTH" AS "SEPALLENGTH" FROM (SELECT ROW_NUMBER() OVER(ORDER BY "SEPALLENGTH") as rn, "SEPALLENGTH" FROM (SELECT * FROM DB2INST1.IRIS)) WHERE rn in(38,39,75,76,112,113))
> (SELECT "SEPALWIDTH" AS "SEPALWIDTH" FROM (SELECT ROW_NUMBER() OVER(ORDER BY "SEPALWIDTH") as rn, "SEPALWIDTH" FROM (SELECT * FROM DB2INST1.IRIS)) WHERE rn in(38,39,75,76,112,113))
> (SELECT "PETALLENGTH" AS "PETALLENGTH" FROM (SELECT ROW_NUMBER() OVER(ORDER BY "PETALLENGTH") as rn, "PETALLENGTH" FROM (SELECT * FROM DB2INST1.IRIS)) WHERE rn in(38,39,75,76,112,113))
> (SELECT "PETALWIDTH" AS "PETALWIDTH" FROM (SELECT ROW_NUMBER() OVER(ORDER BY "PETALWIDTH") as rn, "PETALWIDTH" FROM (SELECT * FROM DB2INST1.IRIS)) WHERE rn in(38,39,75,76,112,113))
> SELECT MAX("ID"), MAX("SEPALLENGTH"), MAX("SEPALWIDTH"), MAX("PETALLENGTH"), MAX("PETALWIDTH") FROM DB2INST1.IRIS
Execution time: 1.8747379779815674 seconds.
Out[27]:
ID
SEPALLENGTH
SEPALWIDTH
PETALLENGTH
PETALWIDTH
count
150.000000
150.000000
150.000000
150.000000
150.000000
mean
75.500000
5.843333
3.054000
3.758667
1.198667
std
43.445368
0.828066
0.433594
1.764420
0.763161
min
1.000000
4.300000
2.000000
1.000000
0.100000
25%
38.500000
5.100000
2.800000
1.600000
0.300000
50%
75.500000
5.800000
3.000000
4.350000
1.300000
75%
112.500000
6.400000
3.300000
5.100000
1.800000
max
150.000000
7.900000
4.400000
6.900000
2.500000
Data manipulation¶
In [28]:
iris = iris[["ID","SEPALLENGTH", "SEPALWIDTH"]]
In [29]:
iris['new'] = iris['SEPALLENGTH'] + iris['SEPALWIDTH'].mean()
> CREATE VIEW "TEMP_VIEW_IRIS_99527_1463749353" AS (SELECT "ID","SEPALLENGTH","SEPALWIDTH" FROM DB2INST1.IRIS)
<< AUTOCOMMIT >>
> DROP VIEW "TEMP_VIEW_IRIS_99527_1463749353"
<< AUTOCOMMIT >>
> SELECT AVG(CAST("SEPALWIDTH" AS FLOAT)) FROM (SELECT "SEPALWIDTH" FROM DB2INST1.IRIS)
Execution time: 0.14851880073547363 seconds.
In [30]:
iris.head()
> CREATE VIEW "TEMP_VIEW_IRIS_42406_1463749360" AS (SELECT "ID","SEPALLENGTH","SEPALWIDTH",("SEPALLENGTH" + 3.054) AS "new" FROM DB2INST1.IRIS)
<< AUTOCOMMIT >>
> DROP VIEW "TEMP_VIEW_IRIS_42406_1463749360"
<< AUTOCOMMIT >>
> SELECT * FROM (SELECT "ID","SEPALLENGTH","SEPALWIDTH",("SEPALLENGTH" + 3.054) AS "new" FROM DB2INST1.IRIS) ORDER BY "ID" ASC FETCH FIRST 5 ROWS ONLY
Out[30]:
ID
SEPALLENGTH
SEPALWIDTH
new
0
1
5.1
3.5
8.154
1
2
4.9
3.0
7.954
2
3
4.7
3.2
7.754
3
4
4.6
3.1
7.654
4
5
5.0
3.6
8.054
Data stays in the database¶
In [31]:
iris
Out[31]:
<ibmdbpy.frame.IdaDataFrame at 0xa3d91d0>
In [32]:
iris.print()
SELECT "ID","SEPALLENGTH","SEPALWIDTH",("SEPALLENGTH" + 3.054) AS "new" FROM DB2INST1.IRIS
Machine Learning¶
- IBM dashDB is more than just a database
- Includes in-database Data Mining algorithms
- Currently available in ibmdbpy :
- K-means, Naive Bayes, Association Rules
- ...
K-means Clustering¶
- Sklearn-like
In [33]:
from ibmdbpy.learn import KMeans
kmeans = KMeans(3) # clustering with 3 clusters
In [34]:
iris = IdaDataFrame(idadb, "IRIS", indexer = "ID")
In [35]:
kmeans.fit_predict(iris).head()
> SELECT COUNT(*) FROM SYSCAT.ROUTINES WHERE ROUTINENAME='KMEANS' AND ROUTINEMODULENAME = 'IDAX'
> CALL IDAX.KMEANS ('idbased=False,model="KMEANS_31778_1463749495",k=3,distance="euclidean",randseed=12345,maxiter=5,intable=IRIS,id="ID"')
<< AUTOCOMMIT >>
<< COMMIT >>
> SELECT * FROM "DB2INST1"."KMEANS_31778_1463749495_MODEL"
> SELECT * FROM "DB2INST1"."KMEANS_31778_1463749495_COLUMNS"
> SELECT * FROM "DB2INST1"."KMEANS_31778_1463749495_COLUMN_STATISTICS"
> SELECT * FROM "DB2INST1"."KMEANS_31778_1463749495_CLUSTERS"
> CALL IDAX.PREDICT_KMEANS ('model="KMEANS_31778_1463749495",outtable="PREDICT_KMEANS_15239_1463749505",intable=IRIS,id="ID"')
<< AUTOCOMMIT >>
<< COMMIT >>
> SELECT distinct TABSCHEMA, TABNAME, OWNER, TYPE from SYSCAT.TABLES WHERE (OWNERTYPE = 'U') ORDER BY "TABSCHEMA","TABNAME"
> SELECT * FROM DB2INST1.PREDICT_KMEANS_15239_1463749505 ORDER BY "ID" ASC FETCH FIRST 5 ROWS ONLY
Out[35]:
ID
CLUSTER_ID
DISTANCE
0
1
1
0.332119
1
2
2
0.228125
2
3
2
0.138505
3
4
2
0.141902
4
5
1
0.382005
In [36]:
kmeans.describe()
KMeans clustering with 3 clusters of sizes 21, 32, 97
Cluster means:
CLUSTERID SEPALLENGTH SEPALWIDTH PETALLENGTH PETALWIDTH CLASS
0 1 5.209375 3.531250 1.671875 0.353125 setosa
1 2 4.695238 3.100000 1.395238 0.190476 setosa
2 3 6.301031 2.886598 4.958763 1.695876 virginica
Within cluster sum of squares by cluster:
[ 2.75907407 22.02570934 170.84912849]
> CALL IDAX.PRINT_MODEL ('model="KMEANS_31778_1463749495"')
<< AUTOCOMMIT >>
Sneak preview¶
- In-database Feature Selection
In [37]:
iris = IdaDataFrame(idadb, "IRIS_DISC", indexer="ID")
In [38]:
from ibmdbpy.feature_selection import info_gain
info_gain(iris)
> SELECT CAST(COUNT(*) AS INTEGER) FROM DB2INST1.IRIS_DISC
> SELECT SUM(-a*LOG(a)) FROM(SELECT COUNT(*) AS a FROM DB2INST1.IRIS_DISC GROUP BY "SEPALLENGTH")
> SELECT SUM(-a*LOG(a)) FROM(SELECT COUNT(*) AS a FROM DB2INST1.IRIS_DISC GROUP BY "SEPALWIDTH")
> SELECT SUM(-a*LOG(a)) FROM(SELECT COUNT(*) AS a FROM DB2INST1.IRIS_DISC GROUP BY "SEPALLENGTH","SEPALWIDTH")
> SELECT SUM(-a*LOG(a)) FROM(SELECT COUNT(*) AS a FROM DB2INST1.IRIS_DISC GROUP BY "PETALLENGTH")
> SELECT SUM(-a*LOG(a)) FROM(SELECT COUNT(*) AS a FROM DB2INST1.IRIS_DISC GROUP BY "SEPALLENGTH","PETALLENGTH")
> SELECT SUM(-a*LOG(a)) FROM(SELECT COUNT(*) AS a FROM DB2INST1.IRIS_DISC GROUP BY "PETALWIDTH")
> SELECT SUM(-a*LOG(a)) FROM(SELECT COUNT(*) AS a FROM DB2INST1.IRIS_DISC GROUP BY "SEPALLENGTH","PETALWIDTH")
> SELECT SUM(-a*LOG(a)) FROM(SELECT COUNT(*) AS a FROM DB2INST1.IRIS_DISC GROUP BY "CLASS")
> SELECT SUM(-a*LOG(a)) FROM(SELECT COUNT(*) AS a FROM DB2INST1.IRIS_DISC GROUP BY "SEPALLENGTH","CLASS")
> SELECT SUM(-a*LOG(a)) FROM(SELECT COUNT(*) AS a FROM DB2INST1.IRIS_DISC GROUP BY "SEPALWIDTH","PETALLENGTH")
> SELECT SUM(-a*LOG(a)) FROM(SELECT COUNT(*) AS a FROM DB2INST1.IRIS_DISC GROUP BY "SEPALWIDTH","PETALWIDTH")
> SELECT SUM(-a*LOG(a)) FROM(SELECT COUNT(*) AS a FROM DB2INST1.IRIS_DISC GROUP BY "SEPALWIDTH","CLASS")
> SELECT SUM(-a*LOG(a)) FROM(SELECT COUNT(*) AS a FROM DB2INST1.IRIS_DISC GROUP BY "PETALLENGTH","PETALWIDTH")
> SELECT SUM(-a*LOG(a)) FROM(SELECT COUNT(*) AS a FROM DB2INST1.IRIS_DISC GROUP BY "PETALLENGTH","CLASS")
> SELECT SUM(-a*LOG(a)) FROM(SELECT COUNT(*) AS a FROM DB2INST1.IRIS_DISC GROUP BY "PETALWIDTH","CLASS")
Out[38]:
SEPALLENGTH
SEPALWIDTH
PETALLENGTH
PETALWIDTH
CLASS
SEPALLENGTH
NaN
0.163915
0.708674
0.675541
0.652284
SEPALWIDTH
0.163915
NaN
0.381823
0.397607
0.376050
PETALLENGTH
0.708674
0.381823
NaN
1.346137
1.356545
PETALWIDTH
0.675541
0.397607
1.346137
NaN
1.378403
CLASS
0.652284
0.376050
1.356545
1.378403
NaN
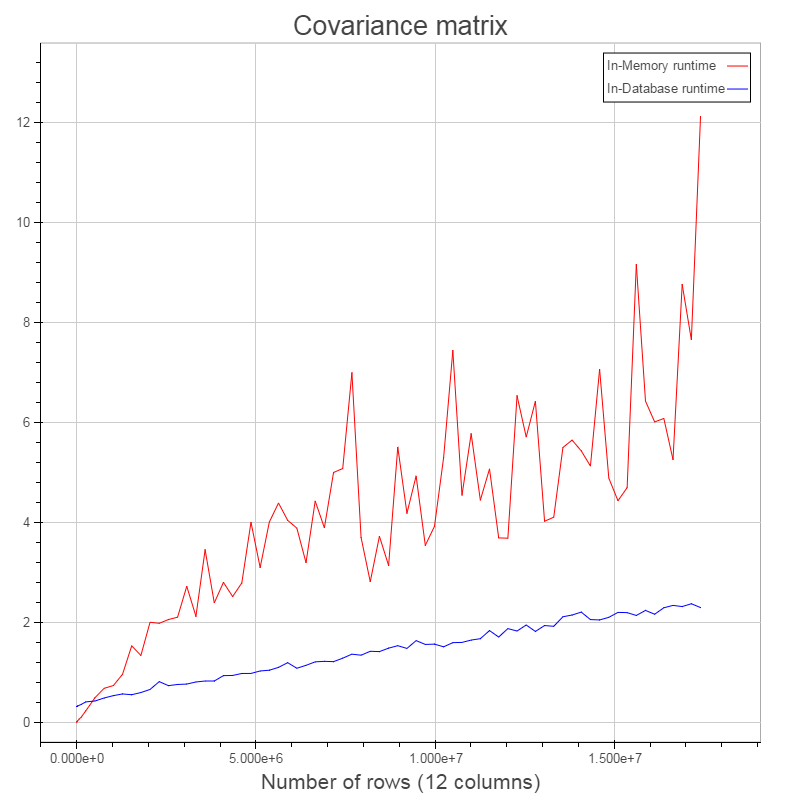
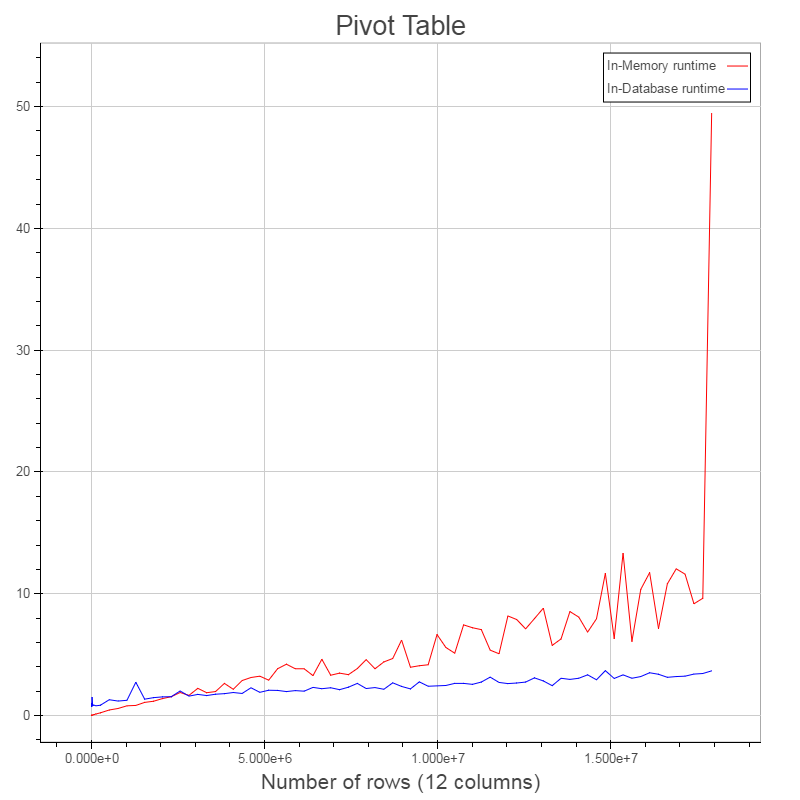
Performance comparison¶
- Comparing in-database and in-memory variant
- in-database: IBM dashDB entry plan on Bluemix
- in-memory: Notebook, i5 2.6GHz - 16GB RAM
Deployment¶
- Distribution via PyPI
pip install ibmdbpy
- Available on GitHub: https://github.com/ibmdbanalytics/ibmdbpy
- License : BSD
Future work¶
- Full test coverage
- More features coming soon:
- Merge, sampling
- More ML wrappers
- In-Database Feature Selection
- In-Database Geospatial Analytics
Conclusion¶
- Ibmdbpy is an interface for in-database computing
- Relies on the database engine
- No data extraction required
- Data freshness
- Security
- Intuitive: Pandas-like, Sklearn-like syntax
- Shows great performance on "big" datasets
- However:
- Requires to connect to a remote database instance
- Designed only for IBM dashDB / IBM DB2
- Covers only a small part of Python analytics capabilities
- Note: For R users, we have a similar interface for R: ibmdbR
Thank you for your attention !¶
Any questions, suggestions ?


In-Database Processing
PyData Berlin 2016
Edouard Fouché
IBM Research & Development
20/05/2016


About me¶

Edouard Fouché¶
- M.Eng. from ESIEE PARIS
- Graduate CS student at Karlsruhe Institute of Technology (KIT)
- Interested in Machine Learning & Cloud Computing
- Working student at IBM since April ’15
- Developing Python tools for in-database analytics
Outline¶
- Motivations
- Python for data science
- Limitations
- Solutions
- Bringing analytics to the data
- IBM dashDB
- Python/SQL
- Ibmdbpy
- Demo !
- Conclusion/Discussion
Motivations¶
- The Python ecosystem for Data Science is rich
- Scipy, Numpy, Pandas
- Scikit-learn
- Matplotlib, Seaborn, Bokeh
- IPython notebooks...

Performance Limitation¶
- CPU, RAM
- Laptop / Workstation

Data Extraction¶
- Data is typically stored in data warehouses
- Big data volumes are impractical to extract

Solutions¶
Out-of-core: Data does not fit in the main memory
- In-Workstation computation, e.g. Dask
- In-Cluster computation, e.g. Hadoop, Spark
- In-Database computation
- Directly in the database engine
In-Database computation¶
Advantageous for several reasons:
- Databases are everywhere !
- Efficiency and scalability
- Data freshness
- Security

- Cloud-based data warehousing system
- Optimized for analytics and data mining
- Integrates BLU technology
- Data compression
- In-memory column store
- Massive Parallel Processing (MPP) scale-out

Task: Compute the mean for each column given the class in the IRIS data set
SELECT AVG(PETALLENGTH), AVG(PETALWIDTH), AVG(SEPALLENGTH), AVG(SEPALWIDTH) FROM IRIS GROUP BY SPECIES
- Only one line in Pandas
In [21]:
from ibmdbpy.sampledata import iris
In [22]:
iris.groupby("species").mean()
Out[22]:
| sepal_length | sepal_width | petal_length | petal_width | |
|---|---|---|---|---|
| species | ||||
| setosa | 5.006 | 3.428 | 1.462 | 0.246 |
| versicolor | 5.936 | 2.770 | 4.260 | 1.326 |
| virginica | 6.588 | 2.974 | 5.552 | 2.026 |

- Pandas-like interface for multiple backends
- Translates Python code into something else (e.g. SQL)
- Supports a lot of backends
However
- Only a subset of functions for each backend
- Cannot make use of backends' specific functions

The SQL-Pushdown approach¶
We translate higher level syntax into SQL
We push them to the underlying database
Everything happens transparently

source : KDnuggets 2015
ibmdbpy¶
- Pandas-like interface for IBM dashDB
- Compatible Python 2.7 up to 3.5
- ODBC or JDBC connection (cross-platform)
- Data analytics
- Database administration

Demo¶
Connect to the database¶
In [23]:
from ibmdbpy import IdaDataBase
idadb = IdaDataBase("DASHDB", verbose=True)
IdaDataBase instances are an abstraction layer for the connection to dashDB
- IDA : In-Database-Analytics
- Database administration
- Database lookup
- Upload Pandas.DataFrame, Download Table
- Drop tables, create views...
Open a pointer to a table¶
In [24]:
from ibmdbpy import IdaDataFrame
iris = IdaDataFrame(idadb, "IRIS", indexer = "ID") # IRIS dataset
> SELECT distinct TABSCHEMA, TABNAME, OWNER, TYPE from SYSCAT.TABLES WHERE (OWNERTYPE = 'U') ORDER BY "TABSCHEMA","TABNAME" > SELECT TRIM(CURRENT_SCHEMA) FROM SYSIBM.SYSDUMMY1
- An IdaDataFrame instance is a pointer to a table in the database
- Data Manipulation
- Non-destructive data manipulation
- Statistics, filtering, sorting...
- Pandas-like syntax
In [25]:
iris.head()
> SELECT * FROM DB2INST1.IRIS ORDER BY "ID" ASC FETCH FIRST 5 ROWS ONLY
Out[25]:
| ID | SEPALLENGTH | SEPALWIDTH | PETALLENGTH | PETALWIDTH | CLASS | |
|---|---|---|---|---|---|---|
| 0 | 1 | 5.1 | 3.5 | 1.4 | 0.2 | setosa |
| 1 | 2 | 4.9 | 3.0 | 1.4 | 0.2 | setosa |
| 2 | 3 | 4.7 | 3.2 | 1.3 | 0.2 | setosa |
| 3 | 4 | 4.6 | 3.1 | 1.5 | 0.2 | setosa |
| 4 | 5 | 5.0 | 3.6 | 1.4 | 0.2 | setosa |
Simple statistics¶
In [26]:
iris.corr()
> SELECT COLNAME, TYPENAME FROM SYSCAT.COLUMNS WHERE TABNAME='IRIS' AND TABSCHEMA='DB2INST1' ORDER BY COLNO
> SELECT CORRELATION("SEPALLENGTH","SEPALWIDTH"), CORRELATION("SEPALLENGTH","PETALLENGTH"), CORRELATION("SEPALLENGTH","PETALWIDTH"), CORRELATION("SEPALWIDTH","PETALLENGTH"), CORRELATION("SEPALWIDTH","PETALWIDTH"), CORRELATION("PETALLENGTH","PETALWIDTH") FROM DB2INST1.IRIS
Execution time: 0.16952085494995117 seconds.
Out[26]:
| SEPALLENGTH | SEPALWIDTH | PETALLENGTH | PETALWIDTH | |
|---|---|---|---|---|
| SEPALLENGTH | 1.000000 | -0.109369 | 0.871754 | 0.817954 |
| SEPALWIDTH | -0.109369 | 1.000000 | -0.420516 | -0.356544 |
| PETALLENGTH | 0.871754 | -0.420516 | 1.000000 | 0.962757 |
| PETALWIDTH | 0.817954 | -0.356544 | 0.962757 | 1.000000 |
In [27]:
iris.describe()
> SELECT COUNT("ID"), COUNT("SEPALLENGTH"), COUNT("SEPALWIDTH"), COUNT("PETALLENGTH"), COUNT("PETALWIDTH") FROM DB2INST1.IRIS
> SELECT AVG(CAST("ID" AS FLOAT)), AVG(CAST("SEPALLENGTH" AS FLOAT)), AVG(CAST("SEPALWIDTH" AS FLOAT)), AVG(CAST("PETALLENGTH" AS FLOAT)), AVG(CAST("PETALWIDTH" AS FLOAT)) FROM DB2INST1.IRIS
> SELECT COUNT("ID"), COUNT("SEPALLENGTH"), COUNT("SEPALWIDTH"), COUNT("PETALLENGTH"), COUNT("PETALWIDTH") FROM DB2INST1.IRIS
> SELECT STDDEV("ID")*(SQRT(150.0)/SQRT(149.0)), STDDEV("SEPALLENGTH")*(SQRT(150.0)/SQRT(149.0)), STDDEV("SEPALWIDTH")*(SQRT(150.0)/SQRT(149.0)), STDDEV("PETALLENGTH")*(SQRT(150.0)/SQRT(149.0)), STDDEV("PETALWIDTH")*(SQRT(150.0)/SQRT(149.0)) FROM DB2INST1.IRIS
> SELECT MIN("ID"), MIN("SEPALLENGTH"), MIN("SEPALWIDTH"), MIN("PETALLENGTH"), MIN("PETALWIDTH") FROM DB2INST1.IRIS
> SELECT * FROM (SELECT COUNT(*) AS "ID" FROM DB2INST1.IRIS WHERE "ID" IS NULL), (SELECT COUNT(*) AS "SEPALLENGTH" FROM DB2INST1.IRIS WHERE "SEPALLENGTH" IS NULL), (SELECT COUNT(*) AS "SEPALWIDTH" FROM DB2INST1.IRIS WHERE "SEPALWIDTH" IS NULL), (SELECT COUNT(*) AS "PETALLENGTH" FROM DB2INST1.IRIS WHERE "PETALLENGTH" IS NULL), (SELECT COUNT(*) AS "PETALWIDTH" FROM DB2INST1.IRIS WHERE "PETALWIDTH" IS NULL)
> SELECT CAST(COUNT(*) AS INTEGER) FROM DB2INST1.IRIS
> (SELECT "ID" AS "ID" FROM (SELECT ROW_NUMBER() OVER(ORDER BY "ID") as rn, "ID" FROM (SELECT * FROM DB2INST1.IRIS)) WHERE rn in(38,39,75,76,112,113))
> (SELECT "SEPALLENGTH" AS "SEPALLENGTH" FROM (SELECT ROW_NUMBER() OVER(ORDER BY "SEPALLENGTH") as rn, "SEPALLENGTH" FROM (SELECT * FROM DB2INST1.IRIS)) WHERE rn in(38,39,75,76,112,113))
> (SELECT "SEPALWIDTH" AS "SEPALWIDTH" FROM (SELECT ROW_NUMBER() OVER(ORDER BY "SEPALWIDTH") as rn, "SEPALWIDTH" FROM (SELECT * FROM DB2INST1.IRIS)) WHERE rn in(38,39,75,76,112,113))
> (SELECT "PETALLENGTH" AS "PETALLENGTH" FROM (SELECT ROW_NUMBER() OVER(ORDER BY "PETALLENGTH") as rn, "PETALLENGTH" FROM (SELECT * FROM DB2INST1.IRIS)) WHERE rn in(38,39,75,76,112,113))
> (SELECT "PETALWIDTH" AS "PETALWIDTH" FROM (SELECT ROW_NUMBER() OVER(ORDER BY "PETALWIDTH") as rn, "PETALWIDTH" FROM (SELECT * FROM DB2INST1.IRIS)) WHERE rn in(38,39,75,76,112,113))
> SELECT MAX("ID"), MAX("SEPALLENGTH"), MAX("SEPALWIDTH"), MAX("PETALLENGTH"), MAX("PETALWIDTH") FROM DB2INST1.IRIS
Execution time: 1.8747379779815674 seconds.
Out[27]:
| ID | SEPALLENGTH | SEPALWIDTH | PETALLENGTH | PETALWIDTH | |
|---|---|---|---|---|---|
| count | 150.000000 | 150.000000 | 150.000000 | 150.000000 | 150.000000 |
| mean | 75.500000 | 5.843333 | 3.054000 | 3.758667 | 1.198667 |
| std | 43.445368 | 0.828066 | 0.433594 | 1.764420 | 0.763161 |
| min | 1.000000 | 4.300000 | 2.000000 | 1.000000 | 0.100000 |
| 25% | 38.500000 | 5.100000 | 2.800000 | 1.600000 | 0.300000 |
| 50% | 75.500000 | 5.800000 | 3.000000 | 4.350000 | 1.300000 |
| 75% | 112.500000 | 6.400000 | 3.300000 | 5.100000 | 1.800000 |
| max | 150.000000 | 7.900000 | 4.400000 | 6.900000 | 2.500000 |
Data manipulation¶
In [28]:
iris = iris[["ID","SEPALLENGTH", "SEPALWIDTH"]]
In [29]:
iris['new'] = iris['SEPALLENGTH'] + iris['SEPALWIDTH'].mean()
> CREATE VIEW "TEMP_VIEW_IRIS_99527_1463749353" AS (SELECT "ID","SEPALLENGTH","SEPALWIDTH" FROM DB2INST1.IRIS)
<< AUTOCOMMIT >>
> DROP VIEW "TEMP_VIEW_IRIS_99527_1463749353"
<< AUTOCOMMIT >>
> SELECT AVG(CAST("SEPALWIDTH" AS FLOAT)) FROM (SELECT "SEPALWIDTH" FROM DB2INST1.IRIS)
Execution time: 0.14851880073547363 seconds.
In [30]:
iris.head()
> CREATE VIEW "TEMP_VIEW_IRIS_42406_1463749360" AS (SELECT "ID","SEPALLENGTH","SEPALWIDTH",("SEPALLENGTH" + 3.054) AS "new" FROM DB2INST1.IRIS)
<< AUTOCOMMIT >>
> DROP VIEW "TEMP_VIEW_IRIS_42406_1463749360"
<< AUTOCOMMIT >>
> SELECT * FROM (SELECT "ID","SEPALLENGTH","SEPALWIDTH",("SEPALLENGTH" + 3.054) AS "new" FROM DB2INST1.IRIS) ORDER BY "ID" ASC FETCH FIRST 5 ROWS ONLY
Out[30]:
| ID | SEPALLENGTH | SEPALWIDTH | new | |
|---|---|---|---|---|
| 0 | 1 | 5.1 | 3.5 | 8.154 |
| 1 | 2 | 4.9 | 3.0 | 7.954 |
| 2 | 3 | 4.7 | 3.2 | 7.754 |
| 3 | 4 | 4.6 | 3.1 | 7.654 |
| 4 | 5 | 5.0 | 3.6 | 8.054 |
Data stays in the database¶
In [31]:
iris
Out[31]:
<ibmdbpy.frame.IdaDataFrame at 0xa3d91d0>
In [32]:
iris.print()
SELECT "ID","SEPALLENGTH","SEPALWIDTH",("SEPALLENGTH" + 3.054) AS "new" FROM DB2INST1.IRIS
Machine Learning¶
- IBM dashDB is more than just a database
- Includes in-database Data Mining algorithms
- Currently available in ibmdbpy :
- K-means, Naive Bayes, Association Rules
- ...
K-means Clustering¶
- Sklearn-like
In [33]:
from ibmdbpy.learn import KMeans
kmeans = KMeans(3) # clustering with 3 clusters
In [34]:
iris = IdaDataFrame(idadb, "IRIS", indexer = "ID")
In [35]:
kmeans.fit_predict(iris).head()
> SELECT COUNT(*) FROM SYSCAT.ROUTINES WHERE ROUTINENAME='KMEANS' AND ROUTINEMODULENAME = 'IDAX'
> CALL IDAX.KMEANS ('idbased=False,model="KMEANS_31778_1463749495",k=3,distance="euclidean",randseed=12345,maxiter=5,intable=IRIS,id="ID"')
<< AUTOCOMMIT >>
<< COMMIT >>
> SELECT * FROM "DB2INST1"."KMEANS_31778_1463749495_MODEL"
> SELECT * FROM "DB2INST1"."KMEANS_31778_1463749495_COLUMNS"
> SELECT * FROM "DB2INST1"."KMEANS_31778_1463749495_COLUMN_STATISTICS"
> SELECT * FROM "DB2INST1"."KMEANS_31778_1463749495_CLUSTERS"
> CALL IDAX.PREDICT_KMEANS ('model="KMEANS_31778_1463749495",outtable="PREDICT_KMEANS_15239_1463749505",intable=IRIS,id="ID"')
<< AUTOCOMMIT >>
<< COMMIT >>
> SELECT distinct TABSCHEMA, TABNAME, OWNER, TYPE from SYSCAT.TABLES WHERE (OWNERTYPE = 'U') ORDER BY "TABSCHEMA","TABNAME"
> SELECT * FROM DB2INST1.PREDICT_KMEANS_15239_1463749505 ORDER BY "ID" ASC FETCH FIRST 5 ROWS ONLY
Out[35]:
| ID | CLUSTER_ID | DISTANCE | |
|---|---|---|---|
| 0 | 1 | 1 | 0.332119 |
| 1 | 2 | 2 | 0.228125 |
| 2 | 3 | 2 | 0.138505 |
| 3 | 4 | 2 | 0.141902 |
| 4 | 5 | 1 | 0.382005 |
In [36]:
kmeans.describe()
KMeans clustering with 3 clusters of sizes 21, 32, 97
Cluster means:
CLUSTERID SEPALLENGTH SEPALWIDTH PETALLENGTH PETALWIDTH CLASS
0 1 5.209375 3.531250 1.671875 0.353125 setosa
1 2 4.695238 3.100000 1.395238 0.190476 setosa
2 3 6.301031 2.886598 4.958763 1.695876 virginica
Within cluster sum of squares by cluster:
[ 2.75907407 22.02570934 170.84912849]
> CALL IDAX.PRINT_MODEL ('model="KMEANS_31778_1463749495"')
<< AUTOCOMMIT >>
Sneak preview¶
- In-database Feature Selection
In [37]:
iris = IdaDataFrame(idadb, "IRIS_DISC", indexer="ID")
In [38]:
from ibmdbpy.feature_selection import info_gain
info_gain(iris)
> SELECT CAST(COUNT(*) AS INTEGER) FROM DB2INST1.IRIS_DISC > SELECT SUM(-a*LOG(a)) FROM(SELECT COUNT(*) AS a FROM DB2INST1.IRIS_DISC GROUP BY "SEPALLENGTH") > SELECT SUM(-a*LOG(a)) FROM(SELECT COUNT(*) AS a FROM DB2INST1.IRIS_DISC GROUP BY "SEPALWIDTH") > SELECT SUM(-a*LOG(a)) FROM(SELECT COUNT(*) AS a FROM DB2INST1.IRIS_DISC GROUP BY "SEPALLENGTH","SEPALWIDTH") > SELECT SUM(-a*LOG(a)) FROM(SELECT COUNT(*) AS a FROM DB2INST1.IRIS_DISC GROUP BY "PETALLENGTH") > SELECT SUM(-a*LOG(a)) FROM(SELECT COUNT(*) AS a FROM DB2INST1.IRIS_DISC GROUP BY "SEPALLENGTH","PETALLENGTH") > SELECT SUM(-a*LOG(a)) FROM(SELECT COUNT(*) AS a FROM DB2INST1.IRIS_DISC GROUP BY "PETALWIDTH") > SELECT SUM(-a*LOG(a)) FROM(SELECT COUNT(*) AS a FROM DB2INST1.IRIS_DISC GROUP BY "SEPALLENGTH","PETALWIDTH") > SELECT SUM(-a*LOG(a)) FROM(SELECT COUNT(*) AS a FROM DB2INST1.IRIS_DISC GROUP BY "CLASS") > SELECT SUM(-a*LOG(a)) FROM(SELECT COUNT(*) AS a FROM DB2INST1.IRIS_DISC GROUP BY "SEPALLENGTH","CLASS") > SELECT SUM(-a*LOG(a)) FROM(SELECT COUNT(*) AS a FROM DB2INST1.IRIS_DISC GROUP BY "SEPALWIDTH","PETALLENGTH") > SELECT SUM(-a*LOG(a)) FROM(SELECT COUNT(*) AS a FROM DB2INST1.IRIS_DISC GROUP BY "SEPALWIDTH","PETALWIDTH") > SELECT SUM(-a*LOG(a)) FROM(SELECT COUNT(*) AS a FROM DB2INST1.IRIS_DISC GROUP BY "SEPALWIDTH","CLASS") > SELECT SUM(-a*LOG(a)) FROM(SELECT COUNT(*) AS a FROM DB2INST1.IRIS_DISC GROUP BY "PETALLENGTH","PETALWIDTH") > SELECT SUM(-a*LOG(a)) FROM(SELECT COUNT(*) AS a FROM DB2INST1.IRIS_DISC GROUP BY "PETALLENGTH","CLASS") > SELECT SUM(-a*LOG(a)) FROM(SELECT COUNT(*) AS a FROM DB2INST1.IRIS_DISC GROUP BY "PETALWIDTH","CLASS")
Out[38]:
| SEPALLENGTH | SEPALWIDTH | PETALLENGTH | PETALWIDTH | CLASS | |
|---|---|---|---|---|---|
| SEPALLENGTH | NaN | 0.163915 | 0.708674 | 0.675541 | 0.652284 |
| SEPALWIDTH | 0.163915 | NaN | 0.381823 | 0.397607 | 0.376050 |
| PETALLENGTH | 0.708674 | 0.381823 | NaN | 1.346137 | 1.356545 |
| PETALWIDTH | 0.675541 | 0.397607 | 1.346137 | NaN | 1.378403 |
| CLASS | 0.652284 | 0.376050 | 1.356545 | 1.378403 | NaN |
Performance comparison¶
- Comparing in-database and in-memory variant
- in-database: IBM dashDB entry plan on Bluemix
- in-memory: Notebook, i5 2.6GHz - 16GB RAM


Deployment¶
- Distribution via PyPI
pip install ibmdbpy
- Available on GitHub: https://github.com/ibmdbanalytics/ibmdbpy
- License : BSD
Future work¶
- Full test coverage
- More features coming soon:
- Merge, sampling
- More ML wrappers
- In-Database Feature Selection
- In-Database Geospatial Analytics
Conclusion¶
- Ibmdbpy is an interface for in-database computing
- Relies on the database engine
- No data extraction required
- Data freshness
- Security
- Intuitive: Pandas-like, Sklearn-like syntax
- Shows great performance on "big" datasets
- However:
- Requires to connect to a remote database instance
- Designed only for IBM dashDB / IBM DB2
- Covers only a small part of Python analytics capabilities
- Note: For R users, we have a similar interface for R: ibmdbR
Thank you for your attention !¶
Any questions, suggestions ?


